-
Posts
97 -
Joined
-
Last visited
About Minh B.
- Birthday 02/04/1982
Custom Fields
-
Gender
Male
-
Location
NOMA NE, Washington, DC
-
Interests
Orchids (National Capital & American Orchid Societies), gardening (National Home Gardening Club) & molecular biology/genetics (American Society for Cell Biology).
Minh B.'s Achievements

Urchin (4/13)
-

Membership Report for Calendar Year 2012
Minh B. replied to Origami's topic in Welcome to WAMAS: FAQ / FYI / Hobby News
That's great news that the org is growing & evidence that the hobby is gaining popularity. Perhaps signs of an economic recovery from an otherwise pricey but rewarding hobby. -
I purchased a glowing green trumpet from him at the last meeting and that thing has nearly doubled in sized. He's great & cuts nice big chunks. I couldn't carry anymore in my box yesterday but definitely buying from his table at the next meeting.
-
She really was great. More testimony that to achieve healthy fish, you need healthy nutrition (from high quality foods such as Jan's Natural food blends) and to minimize stress.
-
Actually translocations (common in cancers) or transposable elements of this size and even bigger (common in many organisms including humans) are quite frequent in eukaryotes. I was just shocked as it generally occurs under selective pressure (i.e. bacteria will 'jump genes' to favor antibiotic resistance, humans will jump genetic material to create new antibodies to fight disease) or a consequence of disease (like cancer). This transposable element phenomena was first reported by Barbara McClintock in maize and she later received a nobel prize for her discovery. The primers part is half true. For the common ciliate, I used for primer #1 and primer #2. For the C. irritans specific set of primers, I used primer #3 and primer #2 because primer #2 is a common primer found in C. irritans as well as all ciliates. Primer #3 is what gives the specificity of C. irrtans. As for the DT result, the difference is also that I used 10 uL of DNA for the PCR instead of 2 uL. Because the DT was so much larger than the QT, I figured I would compensate for the decreased concentration of ich DNA (if there was any) by using 5 times more DNA for PCR...enough to get PCR to work for the DT. If I retry this test using the 5X more DNA from the DT and the same primers as the first test (primers #1 + #2), I should see a 750 bp UNLESS the primer recognition site was abolished due to a mutation/translocation. Yes, this is the same DNA I extracted and used for the first test, and kept frozen until this second test (DNA can be kept indefinitely if frozen and will not go bad). I blasted (http://blast.ncbi.nl...YPE=BlastSearch) primer #3 against the entire genome database (which includes the genome from every sequenced organism) and primer #3 only reported it as C. irritans (with full length, 100% identity). Yes, I can get it sequenced but I plan to redo the nested PCR using triple primers 1 + 2 + 3 so we should get a band at 750 and 75 bp (a double confirmation if you will). I can then cut out the bands of interest and get them sequenced. Would be interesting to see if the ich has 'evolved'. Exactly. There wasn't because the first primer pair gave the expected 750 bp band as the paper reported. The 550 band was supposed to be in this 2nd test with their reported primers, but instead of 550, I got 75.
-
Unfortunately, being that I could not get ciliate rDNA amplification in the first test from my infected DT, I expect using water from a 45-day fish-free tank would be a bigger challenge as I wouldn't be able to normalize against a sample with no bands/PCR. As parasites or living things in general die, one of the first things that occur is the cell and nuclear membranes start to quickly break-down, releasing its contents which will also include DNA in to the surroundings. Once this occurs, bacteria and other opportunistic organisms will start to consume this excess phosphates (from DNA), proteins, etc. A healthy system should have enough good bacteria to break-down all this stuff quickly. Concentration indeed. Please see the new experiment below with DNA extracted from sandbed as well. I extracted DNA from the sandbed, but keep in mind it's been over two weeks since the tank has remained fishless. I took the sand from the front of the tank where it was easily accessible (I'm only 5'2", almost fell in myself), but this is still a great idea. Results below. I used the C. irritans-specific primers this time and I used 5X more template DNA in the PCR reaction than the previous test for broad ciliates. Notice also the amplicon/band is not the expected 550 bp, but much smaller (~75 bp). I assembled the gene databases from two independent labs and I have reason to believe that this strain/variant of C. irritans in my tank had a translocation or moved ~480 bp downstream of the end/reverse primer. I was shocked. Two aligned databases showed single base pair mutations and now I see a large translocation. This parasite is rearranging/mutating within the last 5 years. However, the results do confirm that the DT still had ich a week after being fishless. The second image is the incomplete gene assembly from the two gene databases and the yellow region (consensus sequence with the single bp mutation) is basically missing in this PCR. The blue arrows are the location of the two marine ich-specific primers. Could the mutation(s) be a result of growing fitness or selection (i.e. resistance to drugs used to counter the parasite)? Possibly. Has anyone encountered increased drug resistance when treating their fishes?
-
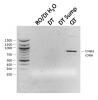
The typical concentration range was 120-170 ng/uL (~120 being the return compartment of the sump). I ordered two sets of primers. This PCR was done nested so I added three primers (two forwards and one common reverse). One pair was not specific to marine ich and is common for the identification of several ciliates and the one extra primer that I added to that pair mix is specific/unique only to marine ich. The PCR from the 3 primers yielded a bright band ~750 (what you see in the image) and a very faint band at ~500 bp, which is the marine ich specific-amplicon. That 500 bp was only observed in the QT but as I didn't take a picture while running the short distance, low intensity bands fade quickly the longer you run the gel to resolve the size difference. I will repeat without the common ciliate primer and only use the marine ich specific primer. But the ciliate primers are only unique to other ciliates so it won't amplify DNA from other organisms -like fish (the paper confirmed this too), snails, crabs, pods, etc., but being that there were no amplicons in the DT or DT sump, it's another testament to how specific these primers are to ciliates. I was absolutely surprised at this result as I predicted the DT to have the strongest intensity. But several factors are against me being 1) the size of the DT (~90 + ~30 gallon sump) vs the QT (30 gallon) which could translate to lower concentration of the parasite in the water per 10 mL; 2) the availability of places that the parasite can adhere to (sand, LR, etc.) that it can't in a QT; 3) a fishless DT vs a QT with a potential pair of hosts. I agree...a negative test would conclude it's indeterminant. Further standardization is required especially for large tanks. Great suggestion! I was just thinking of a place that I could look next to verify the presence of ich in my DT. Never thought of the sandbed. Thanks. I'll give it a try.
-
The PCR assay for the presence of marine ich or Cryptocaryon irritans worked well. Results using the common ciliate pair of primers shows an amplicon of the expected ~750 bp in the quarantine tank (QT). Samples from the display tank (DT) and DT sump, despite being able to extract sufficient DNA, yielded no DNA from the parasite. However, this doesn't conclude that the DT and DT sump are free of the parasite. The DT having live rock (LR) and other non-fish livestock may have likely provided the hiding place for the parasite. This test is good, however, for the detection of the parasite in a QT setup that has nothing but a heater, a pump and a pair of asymptomatic clownfishes that survived the ich war while the others did not (a PBT, foxface and a few anthiases). Some background, the PBT died while in the DT on 1/22, the QT was setup 1/20 but no fishes were transferred until 1/24 (the clown pair and foxface). On 1/26 (two days after transferring to the QT), the foxface died and possibly the culprit for the release of excess parasites into the water column. Clownfish pair still remains asymptomatic to this day. 10 mL of water was collected from both DT sump and QT on 1/31 and the main DT on 2/1, and DNA extracted the same day. PCR was done using primers specfic to the ciliate's ribosomal DNA (rDNA) at 35 cycles with annealing temperature of 50C and extension of 15 sec.
-
+1 I'm sure those coral are resin made and they don't look like realistic growth patterns. I'm still baffled at why the designer would place the aquarium around the fireplace. Clearly the designer has never kept a real aquarium of any kind. And if that was a functional gas or wood burning fireplace, the heat generated would not allow fish keeping unless the owners were looking to make fish soup. Expensive living nowadays doesn't necessarily mean realistic living and there seems to be a sense of pseudo-appreciation for all living things.
-
I took water from my DT and was disappointed to not find any theronts. I proceeded to extract DNA and achieved 170 ng/uL, about the same concentration as the QT. Why a lower concentration in the sump, not quite sure. Perhaps skimmer? But to answer your questions, I used a scope equipped w/polarized light or filters without stain. This works well if there's a high number of speedy theronts that glide on by and catches your attention. I may have to make a stain to see these buggers better. My theronts were on the smaller side, ~15 x 40 um. But I've read their size can vary from one strain to the next, their host species & water temp. Are your scopes equipped with 100x? Maybe test drive your scopes with a drop of pods. They should look like giants on your scopes.
-
Oh, but to answer your questions, I can adjust the sensitivity by decreasing the number of PCR cycles if it turns out the healthy tanks are getting a positive result so that it would be just a very faint band. In which case, this test would be like our pH/alkalinity etc. tests that we do routinely but has different degrees. Sorry, I, like many others I'm sure, are in the binary set of mind where we either have ich or we don't, instead of realizing and seeing your point that perhaps healthy systems have ich but are better equipped to deal with it. Oh, samples I test will remain confidential. I'm not going to blast anyone on the forums if the test turns out positive. I'll treat it like how clinics treat their patients and their personal test results. I'd still show the result for this study but won't say who it came from. So please get me those two samples if anyone has them.
-
Ha, I was thinking the same thing as I was typing the last post. That's why I want to test water from a tank that came from someone who religiously quarantines and their fish show no signs or symptoms. It may be that we are looking for DNA intensity and not the presence of its DNA.
-
The easiest test is to see the theronts under a 100X mag microscope. But after barely seeing theronts in my infected sump and the tomites being extremely hard to see and recognize, this is unreliable. I still think WAMAS should have a microscope for free swimming theronts and strange looking water (algae/bacterial blooms), which I'd be happy to contribute too. It could be a member-only service like the par meter and nowadays, a decent school microscope cost <$400 and I can look into making dyes that stain cell walls. A DNA test is the most conclusive, especially for asymptomatic fish (I feel like I'm Maury Povich offering paternity tests). My clowns have never displayed symptoms on their skin but I'm confident they must have it on their gills. Plus I want to be confident when it's safe to reintroduce fish to the DT and when the parasites are dead in the QT. I need two more samples to test this assay. One from a DT who religiously quarantines all new fish for the ~month period and the fishes have not shown any signs of disease. Another sample would have to come from a fishless DT (no fish at all) for >2 months but still has LR and inverts. I know the second sample may be a challenge to find. I could get the samples at the meeting.
-
Unfortunately, our department is not equipped with video capture for a regular light microscope. Instead, we have two $1.5M (that's right...million) fluorescent scopes that are and several regular light scopes equipped with 100X. The one scope that has a camera has a max 40X lens. I'll take a pic w/the 40X but it won't look as convincing as with a 100X and unfortunately, you won't be able to see the theronts being the road runner. Yes, we still use detergents. Easy & less messy than liquid nitrogen. You're right...I did not sterilize the QT. Yes, I have access to almost all research journals as our National Library of Medicine is on campus. And NLM isn't just for medicine, but all general/basic research as well. Let me know if you can't access a paper and I'll get you a PDF. There's actually many research articles on marine ich. Some, to be honest, are quite bad. I came across one that proposed an 'alternative' form of treatment but their study showed ich numbers came back with the vengeance after 3 weeks.
-
Jan, is it ok if your tank has an explosion or possibly too much stomatellas?
-
Thanks. I will do that. Short answer: 7 days after introducing fish to the QT is plenty! However, it's actually quite easy to spot the theronts visually with a 100X microscope. These things are like race cars...they zoom by so quickly you can't keep up! But if you look carefully enough, you'll find their surface is riddled with cilia/hair-like structures. Long answer: I was very curious about this too. Today, I took to work, 10 mL of water from my DT's return pump compartment of the sump/refugium and 10 mL of water from the QT with the 2 clownfishes that survived. I first checked on the microscope at 100X and saw the highest concentration of theronts (free-swimming infective stage), tomites (very difficult to see) and what may look to be tomonts as well in the QT. I was quite shocked to see this, but to answer your question, the QT was setup exactly a week ago today (last Thursday) and already there were so much more activity than in the return compartment of my sump. It possible the fishes that died with lots of trophonts in the QT released a large amount of protomonts after the fishes died in an attempt to find a new host. I'm not familiar with the parasites response after their host dies. If anyone knows, please chime in. However, I saw very few theronts in the sump suggesting they're starting to die off, but there were still many tomites. Tomorrow, I will grab some water from the actual DT itself where the fishes were housed. Because I saw more activity in the QT than the DT's sump, my prediction is that I should get a higher DNA yield in the QT. Although the paper used repetitive freeze/thaw cycles to burst open the parasite, I came up with a protocol that combines DNA extraction from both bacteria and human cells, and proceeded to extract both 10 mL samples. Briefly, samples were spun down at 10,000 rpm's for 10 min, lysed with a strong detergent and DNA precipitated with phenol (high toxic), chloroform and then ethanol. The DNA was solubilized and quantified by UV and guess what? QT's DNA concentration was 165 ng/uL and DT return sump's concentration was 117 ng/uL. The paper claims that 45 pg of DNA is the minimum for the assay, and assuming the majority of the DNA I extracted is from the parasite, I have >2000X the necessary amount of DNA for the assay from just 10 mL of sample. The real test is when I get the primers next week to run the assay.